PatchLabelSampler#
from matplotlib import pyplot as plt
from wholeslidedata import WholeSlideAnnotation
from wholeslidedata.visualization.plotting import plot_annotations, plot_boxes
Classification#
See also
The Camelyon Example in which a classification task is done.
from wholeslidedata.samplers.patchlabelsampler import ClassificationPatchLabelSampler
wsa = WholeSlideAnnotation('/tmp/TCGA-21-5784-01Z-00-DX1.xml')
annotation = wsa.annotations[0]
classification_patch_label_sampler = ClassificationPatchLabelSampler(123)
classification_label = classification_patch_label_sampler.sample(wsa=wsa, point=annotation.center, size=None, ratio=1)
print(wsa.annotations[0].label.value, classification_label)
1 [1]
Segmentation#
See also
The BACH Example in which a segmentation task is done.
from wholeslidedata.samplers.patchlabelsampler import SegmentationPatchLabelSampler
wsa = WholeSlideAnnotation('/tmp/TCGA-21-5784-01Z-00-DX1.xml')
annotation = wsa.annotations[0]
segmentation_patch_label_sampler= SegmentationPatchLabelSampler()
segmentation_label = patch_label_sampler.sample(wsa=wsa, point=annotation.center, size=annotation.size, ratio=1)
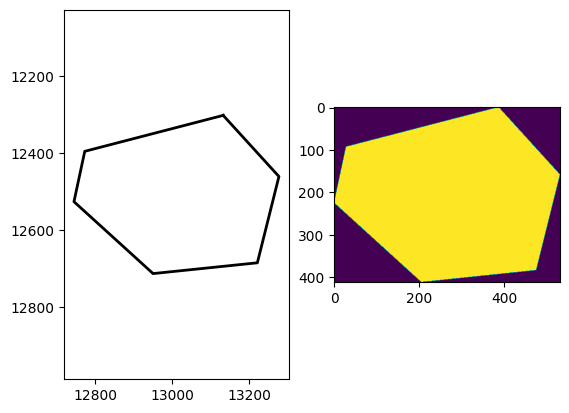
fig, axes = plt.subplots(1,2)
plot_annotations([annotation], ax=axes[0])
axes[0].invert_yaxis()
axes[1].imshow(segmentation_label)
plt.show()
Detection#
See also
The TIGER Example in which a detection task is done with the same data as used below
!aws s3 cp --no-sign-request s3://tiger-training/wsirois/wsi-level-annotations/images/100B.tif /tmp
!aws s3 cp --no-sign-request s3://tiger-training/wsirois/wsi-level-annotations/annotations-tissue-cells-xmls/100B.xml /tmp
download: s3://tiger-training/wsirois/wsi-level-annotations/images/100B.tif to ../../../../../../../../../../../tmp/100B.tif
download: s3://tiger-training/wsirois/wsi-level-annotations/annotations-tissue-cells-xmls/100B.xml to ../../../../../../../../../../../tmp/100B.xml
wsa = WholeSlideAnnotation('/tmp/100B.xml', sample_label_names=['roi'])
roi = wsa.sampling_annotations[0]
from wholeslidedata.samplers.patchlabelsampler import DetectionPatchLabelSampler
detection_patch_sampler = DetectionPatchLabelSampler(max_number_objects=100, detection_labels='lymphocytes and plasma cells')
objects = np.array(detection_patch_sampler.sample(wsa, roi.center, roi.size, ratio=1))
objects[objects[...,-1]>0].astype("int32")
array([[ 248, 89, 266, 107, 6, 1],
[ 265, 867, 283, 885, 6, 1],
[ 340, 343, 358, 361, 6, 1],
[ 390, 766, 408, 784, 6, 1],
[ 394, 792, 412, 810, 6, 1],
[ 420, 353, 438, 371, 6, 1],
[ 425, 331, 443, 349, 6, 1],
[ 433, 319, 451, 337, 6, 1],
[ 436, 1117, 454, 1135, 6, 1],
[ 439, 336, 457, 354, 6, 1],
[ 452, 308, 470, 326, 6, 1],
[ 483, 375, 501, 393, 6, 1],
[ 488, 1133, 506, 1151, 6, 1],
[ 489, 405, 507, 423, 6, 1],
[ 570, 408, 588, 426, 6, 1],
[ 604, 673, 622, 691, 6, 1],
[ 617, 501, 635, 519, 6, 1],
[ 630, 459, 648, 477, 6, 1],
[ 648, 341, 666, 359, 6, 1],
[ 667, 468, 685, 486, 6, 1],
[ 744, 331, 762, 349, 6, 1],
[ 754, 709, 772, 727, 6, 1],
[ 770, 331, 788, 349, 6, 1],
[ 805, 724, 823, 742, 6, 1]], dtype=int32)